When analyzing LC/GC-MS data, it may be necessary to align your chromatogram with the analytical trace so that the peaks match across the different spectra.
For batch and real-time analyses with Mgears, such alignment can be calculated and applied automatically for each sample by the software, or otherwise introduced manually by the user during analysis setup then applied to all samples automatically.
Manual alignment may be recommended in some cases. The below describes a quick way to calculate the time-shift required to align chromatograms with Mnova.
View the traces that are available for your data
Use the Mnova Mass Browser tool to view the traces that are available in your dataset. To do so, you must open your input data by clicking on File > Open and select one of your input data, or simply drag and drop your data file into Mnova.
Then, click on the Show Mass Browser option in the Mass Tools section.
A window with the input spectra and another with MS Browser information will open. Here, you will be able to see which Functions and Traces are available for your dataset (for more information about the MS Browser and available traces, please refer to the Mnova manual).
Calculate the time-shift correction
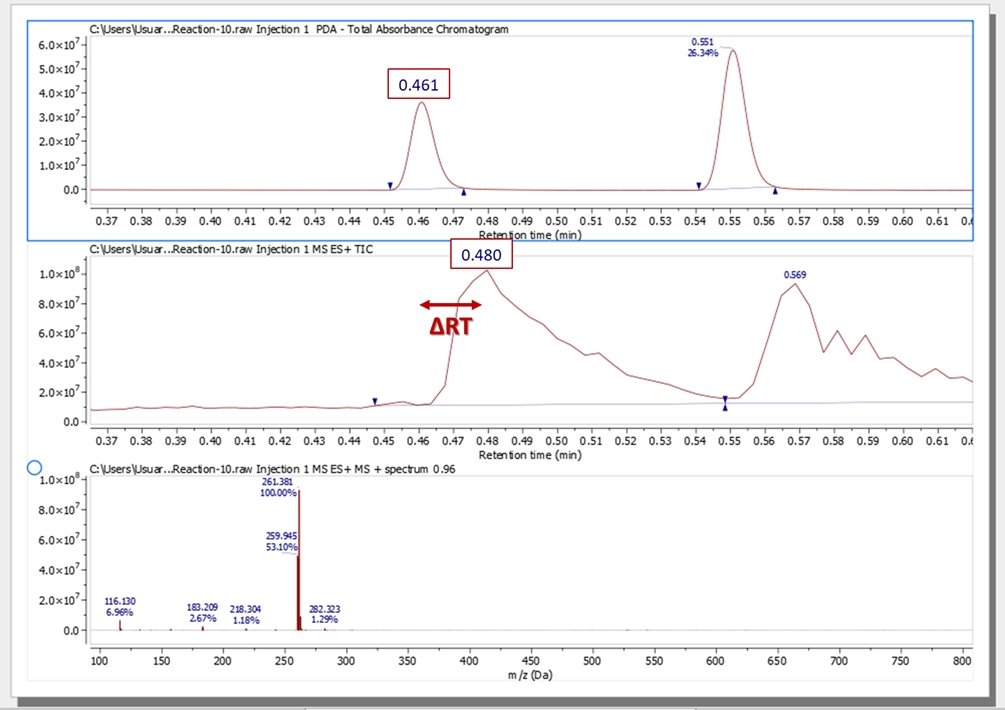
Identify a peak on the Total Absorbance Chromatogram and the MS ES+TIC and annotate its chemical shift for each of the chromatograms.
Subtract these two numbers to obtain the time-shift correction (ΔRT):
ΔRT = RTTIC - RTUV = 0.019
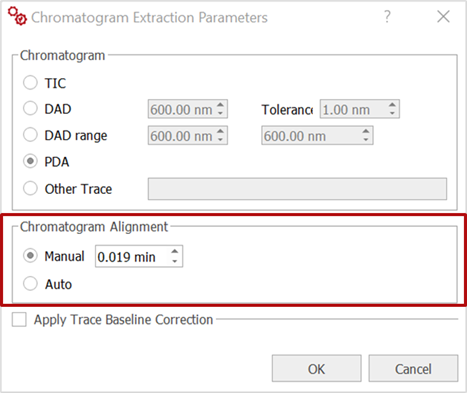
Introduce the time-shift correction into Mgears plugins
Add the ΔRT value to the Manual alignment field in the Gears plugin settings.
It is important to note here that when retention time (RT) values are declared and used in the analysis to match compounds (such as with the Chrom Reaction Optimization analysis), those declared values can proceed from a previously aligned chromatogram or from an unaligned chromatogram.
If the declared RTs proceed from an aligned chromatogram, then once the time-shift correction (ΔRT) is applied to the analyzed chromatograms, the compounds’ matching will be successfully achieved.
If the declared RTs correspond to an unaligned chromatogram, these values need to be corrected in a similar manner to the analyzed chromatograms by applying the time-shift correction. Otherwise, compound matching will fail. For such cases, an RT alignment correction option is available in the plugin settings and must be enabled in order to obtain correct results.
The results spectra will be correctly aligned.
Check out Chrom Cal and Chrom Reaction Optimization pages.