(Last Updated on 25th Jan 2022)
Mnova Verify is an Mnova plugin, that is fully integrated into the standard Mnova architecture, and that benefits from interaction with all other plugins.
It’s a tool designed for synthetic and medicinal chemists, which automatically checks a proposed structure against a set of analytical data, LCMS or GCMS and/or 1D NMR and/or 2D NMR, as well as full structure assignment to NMR and MS data.
In full automation mode, Verify does the same checks that a chemist would carry out to ensure their compound has the expected structure using MS and NMR data.
Verifications in Single Mode
Check your spectra against a proposed molecular structure in just a few steps.
1- LOAD AND PROCESS YOUR FILES
The first thing you should do is to load your spectra dataset and draw or load your molecular structure(s):
Use Mnova Molecule Editor to draw the appropriate structure(s) or load them via a compatible file format (.mol, .sdf, etc.)
Finally, process your spectra using the usual processing tools such as Phase and Baseline Correction, and Apodization or apply any 'Advised Processing'.
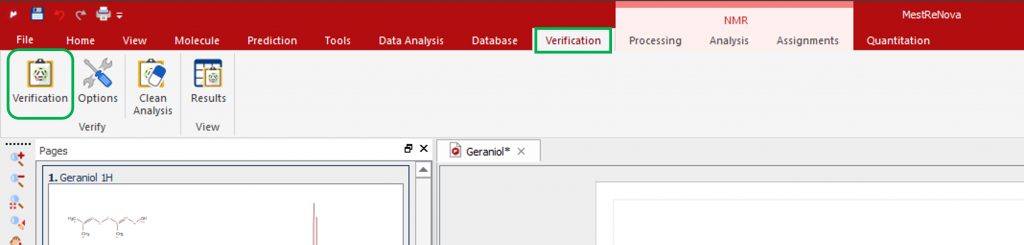
2- VERIFICATION SETUP
On the Verification menu, click on the Verification button to open the Verification Setup window:
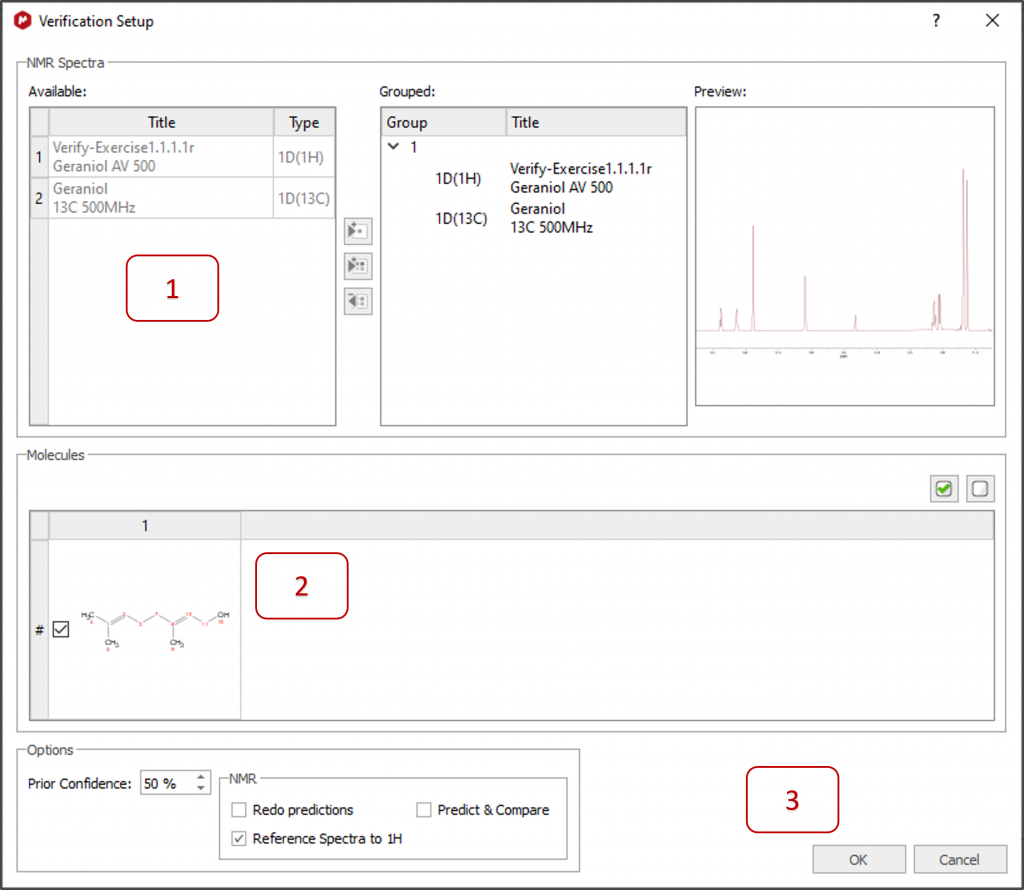
1 - All available spectra are automatically selected for the verification. Move spectra from left to right if you want them to be ignored or to group them differently. A preview for each spectrum is displayed on the right.
2- All available molecules are automatically selected. Select only one for a single verification or several to compare and rank them via Mnova Verify.
3- Select your preferences in 'Options' and click OK
3 – RESULTS AND ANALYSIS
Mnova Verify will provide you a multiplet analysis, solvent identification and verification results:
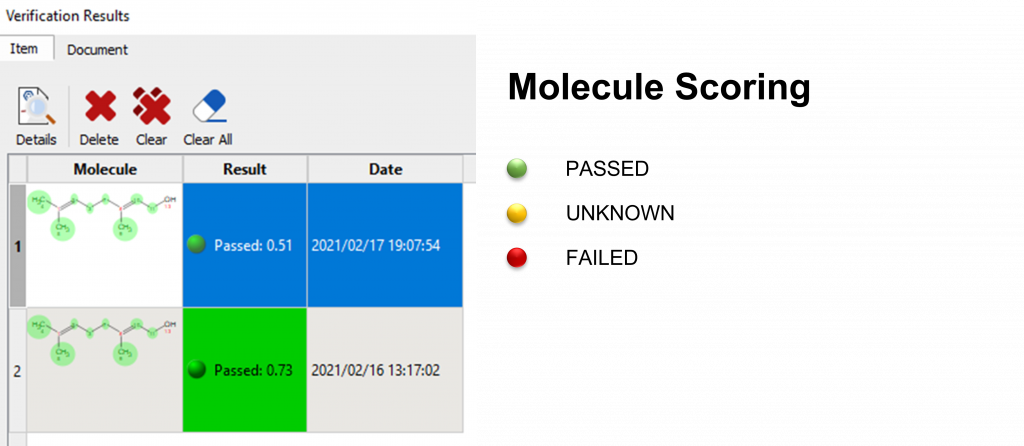
The Verification Results Panel now displays the following information:
Overview - Result, Purity, Date and Verification options.
Spectra Quality - Problems detected concerning the quality of the experimental data.
Diagnostics - Warning messages.
Tests - Score, Significance and Quality values.
The color coded atom assignment is represented as follows:
Finally, the color coding for the Molecule Results is as follows::
Verifications in Batch Mode
Mnova Verify can also be integrated into a fully automated workflow in which data processing, analysis and result reporting and archiving is handled entirely by Mnova's automation engine Mnova Gears. Such a workflow can run in batch or real-time modes and is extremely valuable in QC environments where libraries of compounds are regularly checked. You can use the following link to learn more about Mnova Gears Verify